PCR Primer Design Guidelines
时间:2018年09月21日 08:52:35点击:2014类别:技术开发
PCR Primer Design Guidelines
PCR (Polymerase Chain Reaction)
Polymerase Chain Reaction is widely held as one of the most important inventions of the 20th century in molecular biology. Small amounts of the genetic material can now be amplified to be able to a identify, manipulate DNA, detect infectious organisms, including the viruses that cause AIDS, hepatitis, tuberculosis, detect genetic variations, including mutations, in human genes and numerous other tasks.
PCR involves the following three steps: Denaturation, Annealing and Extension. First, the genetic material is denatured, converting the double stranded DNA molecules to single strands. The primers are then annealed to the complementary regions of the single stranded molecules. In the third step, they are extended by the action of the DNA polymerase. All these steps are temperature sensitive and the common choice of temperatures is 94oC, 60oC and 70oC respectively. Good primer design is essential for successful reactions. The important design considerations described below are a key to specific amplification with high yield. The preferred values indicated are built into all our products by default.
1. Primer Length: It is generally accepted that the optimal length of PCR primers is 18-22 bp. This length is long enough for adequate specificity and short enough for primers to bind easily to the template at the annealing temperature.
2. Primer Melting Temperature: Primer Melting Temperature (Tm) by definition is the temperature at which one half of the DNA duplex will dissociate to become single stranded and indicates the duplex stability. Primers with melting temperatures in the range of 52-58 oC generally produce the best results. Primers with melting temperatures above 65oC have a tendency for secondary annealing. The GC content of the sequence gives a fair indication of the primer Tm. All our products calculate it using the nearest neighbor thermodynamic theory, accepted as a much superior method for estimating it, which is considered the most recent and best available.
Formula for primer Tm calculation:
Melting Temperature Tm(K)={ΔH/ ΔS + R ln(C)}, Or Melting Temperature Tm(oC) = {ΔH/ ΔS + R ln(C)} - 273.15 where
ΔH (kcal/mole) : H is the Enthalpy. Enthalpy is the amount of heat energy possessed by substances. ΔH is the change in Enthalpy. In the above formula the ΔH is obtained by adding up all the di-nucleotide pairs enthalpy values of each nearest neighbor base pair.
ΔS (kcal/mole) : S is the amount of disorder a system exhibits is called entropy. ΔS is change in Entropy. Here it is obtained by adding up all the di-nucleotide pairs entropy values of each nearest neighbor base pair. An additional salt correction is added as the Nearest Neighbor parameters were obtained from DNA melting studies conducted in 1M Na+ buffer and this is the default condition used for all calculations.
ΔS (salt correction) = ΔS (1M NaCl )+ 0.368 x N x ln([Na+])
Where
N is the number of nucleotide pairs in the primer ( primer length -1).
[Na+] is salt equivalent in mM.
[Na+] calculation:
[Na+] = Monovalent ion concentration +4 x free Mg2+.
3. Primer Annealing Temperature: The primer melting temperature is the estimate of the DNA-DNA hybrid stability and critical in determining the annealing temperature. Too high Ta will produce insufficient primer-template hybridization resulting in low PCR product yield. Too low Ta may possibly lead to non-specific products caused by a high number of base pair mismatches,. Mismatch tolerance is found to have the strongest influence on PCR specificity.
Ta = 0.3 x Tm(primer) + 0.7 Tm (product) – 14.9
where,
Tm(primer) = Melting Temperature of the primers
Tm(product) = Melting temperature of the product
4. GC Content: The GC content (the number of G's and C's in the primer as a percentage of the total bases) of primer should be 40-60%.
5. GC Clamp: The presence of G or C bases within the last five bases from the 3' end of primers (GC clamp) helps promote specific binding at the 3' end due to the stronger bonding of G and C bases. More than 3 G's or C's should be avoided in the last 5 bases at the 3' end of the primer.
6. Primer Secondary Structures: Presence of the primer secondary structures produced by intermolecular or intramolecular interactions can lead to poor or no yield of the product. They adversely affect primer template annealing and thus the amplification. They greatly reduce the availability of primers to the reaction.
i) Hairpins: It is formed by intramolecular interaction within the primer and should be avoided. Optimally a 3' end hairpin with a ΔG of -2 kcal/mol and an internal hairpin with a ΔG of -3 kcal/mol is tolerated generally.
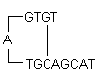
ΔG definition: The Gibbs Free Energy G is the measure of the amount of work that can be extracted from a process operating at a constant pressure. It is the measure of the spontaneity of the reaction. The stability of hairpin is commonly represented by its ΔG value, the energy required to break the secondary structure. Larger negative value for ΔG indicates stable, undesirable hairpins. Presence of hairpins at the 3' end most adversely affects the reaction.
ΔG = ΔH – TΔS
ii) Self Dimer: A primer self-dimer is formed by intermolecular interactions between the two (same sense) primers, where the primer is homologous to itself. Generally a large amount of primers are used in PCR compared to the amount of target gene. When primers form intermolecular dimers much more readily than hybridizing to target DNA, they reduce the product yield. Optimally a 3' end self dimer with a ΔG of -5 kcal/mol and an internal self dimer with a ΔG of -6 kcal/mol is tolerated generally.
iii) Cross Dimer: Primer cross dimers are formed by intermolecular interaction between sense and antisense primers, where they are homologous. Optimally a 3' end cross dimer with a ΔG of -5 kcal/mol and an internal cross dimer with a ΔG of -6 kcal/mol is tolerated generally.
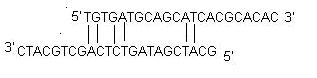
7. Repeats: A repeat is a di-nucleotide occurring many times consecutively and should be avoided because they can misprime. For example: ATATATAT. A maximum number of di-nucleotide repeats acceptable in an oligo is 4 di-nucleotides.
8. Runs: Primers with long runs of a single base should generally be avoided as they can misprime. For example, AGCGGGGGATGGGG has runs of base 'G' of value 5 and 4. A maximum number of runs accepted is 4bp.
9. 3' End Stability: It is the maximum ΔG value of the five bases from the 3' end. An unstable 3' end (less negative ΔG) will result in less false priming.
10. Avoid Template Secondary Structure: A single stranded Nucleic acid sequences is highly unstable and fold into conformations (secondary structures). The stability of these template secondary structures depends largely on their free energy and melting temperatures(Tm). Consideration of template secondary structures is important in designing primers, especially in qPCR. If primers are designed on a secondary structures which is stable even above the annealing temperatures, the primers are unable to bind to the template and the yield of PCR product is significantly affected. Hence, it is important to design primers in the regions of the templates that do not form stable secondary structures during the PCR reaction. Our products determine the secondary structures of the template and design primers avoiding them.
11. Avoid Cross Homology: To improve specificity of the primers it is necessary to avoid regions of homology. Primers designed for a sequence must not amplify other genes in the mixture. Commonly, primers are designed and then BLASTed to test the specificity. Our products offer a better alternative. You can avoid regions of cross homology while designing primers. You can BLAST the templates against the appropriate non-redundant database and the software will interpret the results. It will identify regions significant cross homologies in each template and avoid them during primer search.
Parameters for Primer Pair Design
1. Amplicon Length: The amplicon length is dictated by the experimental goals. For qPCR, the target length is closer to 100 bp and for standard PCR, it is near 500 bp. If you know the positions of each primer with respect to the template, the product is calculated as: Product length = (Position of antisense primer-Position of sense primer) + 1.
2. Product Position: Primer can be located near the 5' end, the 3' end or any where within specified length. Generally, the sequence close to the 3' end is known with greater confidence and hence preferred most frequently.
3. Tm of Product: Melting Temperature (Tm) is the temperature at which one half of the DNA duplex will dissociate and become single stranded. The stability of the primer-template DNA duplex can be measured by the melting temperature (Tm).
4. Optimum Annealing Temperature (Ta Opt): The formula of Rychlik is most respected. Our products use this formula to calculate it and thousands of our customers have reported good results using it for the annealing step of the PCR cycle. It usually results in good PCR product yield with minimum false product production.
Ta Opt = 0.3 x(Tm of primer) + 0.7 x(Tm of product) - 14.9
where
Tm of primer is the melting temperature of the less stable primer-template pair
Tm of product is the melting temperature of the PCR product.
5. Primer Pair Tm Mismatch Calculation: The two primers of a primer pair should have closely matched melting temperatures for maximizing PCR product yield. The difference of 5oC or more can lead no amplification.
Primer Design using Software
A number of primer design tools are available that can assist in PCR primer design for new and experienced users alike. These tools may reduce the cost and time involved in experimentation by lowering the chances of failed experimentation.
Primer Premier follows all the guidelines specified for PCR primer design. Primer Premier can be used to design primers for single templates, alignments, degenerate primer design, restriction enzyme analysis. contig analysis and design of sequencing primers.
The guidelines for qPCR primer design vary slightly. Software such as AlleleID and Beacon Designer can design primers and oligonucleotide probes for complex detection assays such as multiplex assays, cross species primer design, species specific primer design and primer design to reduce the cost of experimentation.
PrimerPlex is a software that can design primers for Multiplex PCR and multiplex SNP genotyping assays.
楼主ヾ(。`Д´。)
喜欢:(2014) 回复:(2)
以下为回复内容
读后有收获可以添加作者微信共同交流

1#楼的root于2018年09月21日 22:32:30回复道:
千万别以为只有什么平面设计行业才会玩Mac,下面是一些本系统会经常用到的。
0. 4Peaks,序列查看,Blast。
1. EnzymeX,序列酶切位点分析,双酶切Buffer选择等。
3. SerialCloner,序列分析。
4. MacVector,序列分析。
5. Primer-Blast,引物设计。
6. Graphpad Prism,数据分析与作图。
3. Mathematica,高级计算器。
7. iWork, nothing to say。
8. Library.nu,就是以前的gigapedia,盗版书集散地。
9. Nature News和Bioon.com,生物研究资讯网站。
10. Biodiscover.com,生物探索,中文,所谓的Web2.0生物聚合门户。
11. Biocompare.com和Labome.com,生物试剂、抗体、蛋白、siRNA等的购买导航网站,尤其是后者中的一些试剂Review,非常实用。
13. Papers,Endnote,文献管理。
20. Red Alert,Spore,大家都是要休息的嘛。
2#楼的九九六等福报于2024年04月18日 15:16:37回复道:
PCR引物设计指南
PCR(聚合酶链反应)
聚合酶链反应被广泛认为是20世纪分子生物学中最重要的发明之一。现在可以扩增少量的遗传物质,以便能够识别、操纵DNA、检测传染性生物体,包括导致艾滋病、肝炎、结核病的病毒,检测人类基因中的遗传变异,包括突变和许多其他任务。
PCR包括以下三个步骤:变性、退火和延伸。首先,遗传物质变性,将双链DNA分子转化为单链。然后将引物退火到单链分子的互补区域。在第三步中,它们通过DNA聚合酶的作用而延伸。所有这些步骤都对温度敏感,通常选择的温度是 94oC, 60oC 和 70oC。良好的引物设计对于成功的反应至关重要。下面描述的重要设计考虑因素是实现高产量特定扩增的关键。默认情况下,指示的首选值内置于我们所有产品中。
1.引物长度:人们普遍认为PCR引物的最佳长度为18-22 bp。该长度足够长,具有足够的特异性,并且足够短,引物在退火温度下很容易与模板结合。
2.引物熔化温度:引物熔化温度(Tm)的定义是DNA双链体的一半解离成为单链的温度,表明双链体的稳定性。熔化温度在52-58范围内的底漆oC 通常产生最佳结果。熔化温度高于 65 的底漆oC 有二次退火的倾向。序列的GC含量给出了引物T的公平指示m.我们所有的产品都使用最近邻热力学理论来计算它,该理论被认为是一种更好的估计方法,被认为是最新和最好的方法。
底漆T的配方m计算:
熔融温度 Tm(K)={ΔH/ ΔS + R ln(C)},或熔融温度 Tm(oC) = {ΔH/ ΔS + R ln(C)} - 273.15 其中
ΔH (kcal/mole) : H 是焓。焓是物质所拥有的热能。ΔH是焓的变化。在上式中,ΔH是通过将每个最近邻碱基对的所有双核苷酸对焓值相加而获得的。
ΔS(千卡/摩尔):S 是系统表现出的无序量,称为熵。ΔS 是熵的变化。这里是通过将每个最近邻碱基对的所有双核苷酸对熵值相加而获得的。由于最近邻参数是从在 1M Na+ 缓冲液中进行的 DNA 熔解研究中获得的,因此添加了额外的盐校正,这是用于所有计算的默认条件。
ΔS(盐校正)= ΔS (1M NaCl )+ 0.368 x N x ln([Na+])
其中
N 是引物中核苷酸对的数量(引物长度 -1)。
[Na+] 是以 mM 为单位的盐当量。
[Na+] 计算:
[Na+] = 一价离子浓度 +4 x 游离 Mg2+。
3. 引物退火温度:引物熔化温度是 DNA-DNA 杂交稳定性的估计值,对确定退火温度至关重要。T 过高一个将产生不充分的引物-模板杂交,导致PCR产物产物产量低。T 过低一个可能导致由大量碱基对错配引起的非特异性产物。研究发现,错配耐受性对PCR特异性的影响最大。
T一个= 0.3 x 吨m(底漆)+ 0.7 Tm(产品)– 14.9
其中,
Tm(引物) = 引物的熔化温度
Tm(产品)=产品的熔点温度
4.GC含量:引物的GC含量(引物中G和C的数量占总碱基的百分比)应为40-60%。
5. GC 钳:由于 G 和 C 碱的结合更强,引物 3' 端(GC 钳)的最后五个碱基内存在 G 或 C 碱基,有助于促进 3' 端的特异性结合。在引物 3' 末端的最后 5 个碱基中应避免超过 3 个 G 或 C。
6. 引物二级结构:分子间或分子内相互作用产生的引物二级结构的存在会导致产物收率低或没有产率。它们会对引物模板退火产生不利影响,从而对扩增产生不利影响。它们大大降低了引物对反应的可用性。
i) 发夹:它是由引物内的分子内相互作用形成的,应避免使用。通常,通常可以耐受 ΔG 为 -2 kcal/mol 的 3' 端发夹和 ΔG 为 -3 kcal/mol 的内发夹。

ΔG 定义:吉布斯自由能 G 是从恒定压力下运行的过程中可以提取的功量的量度。它是反应自发性的量度。发夹的稳定性通常由其ΔG值表示,即破坏二级结构所需的能量。ΔG 的负值越大,表示发夹稳定、不需要。3'端发夹的存在对反应产生最不利的影响。
ΔG = ΔH – TΔS
ii) 自二聚体:引物自二聚体由两个(相同义)引物之间的分子间相互作用形成,其中引物与自身同源。通常,与靶基因的数量相比,PCR中使用了大量的引物。当引物形成分子间二聚体比与靶DNA杂交更容易时,它们会降低产物产率。理想情况下,通常耐受 ΔG 为 -5 kcal/mol 的 3' 端自二聚体和 ΔG 为 -6 kcal/mol 的内部自二聚体。
iii) 交叉二聚体:引物交叉二聚体由正义引物和反义引物之间的分子间相互作用形成,它们是同源的。理想情况下,通常可耐受 ΔG 为 -5 kcal/mol 的 3' 端交叉二聚体和 ΔG 为 -6 kcal/mol 的内部交叉二聚体。
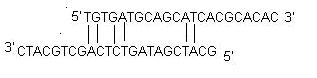
7. 重复:重复是连续多次出现的双核苷酸,应避免,因为它们可能会误导。例如:ATATATAT。寡核苷酸中可接受的最大双核苷酸重复次数为 4 个双核苷酸。
8. 运行:通常应避免使用单个碱基的长运行引物,因为它们可能会产生错误。例如,AGCGGGGGATGGGG 具有值 5 和 4 的基数“G”的运行。接受的最大运行次数为 4bp。
9. 3' 端稳定性:它是从 3' 端开始的五个碱基的最大 ΔG 值。不稳定的 3' 端(负 ΔG 较小)将导致较少的假启动。
10.避免模板二级结构:单链核酸序列高度不稳定,并折叠成构象(二级结构)。这些模板二级结构的稳定性很大程度上取决于其自由能和熔融温度(Tm).在设计引物时,考虑模板二级结构非常重要,尤其是在qPCR中。如果引物设计在二级结构上,即使在退火温度以上也能稳定,则引物无法与模板结合,并且PCR产物的产量会受到显着影响。因此,在PCR反应过程中不形成稳定二级结构的模板区域设计引物非常重要。我们的产品确定模板的二级结构,并设计避免它们的引物。
11.避免交叉同源性:为了提高引物的特异性,有必要避免同源性区域。为序列设计的引物不得扩增混合物中的其他基因。通常,先设计引物,然后进行爆破以测试特异性。我们的产品提供了更好的选择。在设计引物时,可以避免交叉同源区域。您可以针对适当的非冗余数据库对模板进行爆破,软件将解释结果。它将识别每个模板中具有显着交叉同源性的区域,并在引物搜索过程中避免它们。
引物对设计的参数
1. 扩增子长度:扩增子长度由实验目标决定。对于qPCR,目标长度接近100 bp,对于标准PCR,目标长度接近500 bp。如果您知道每个引物相对于模板的位置,则乘积的计算公式为:产品长度 =(反义引物的位置-正义引物的位置)+ 1。
2.产品位置:底漆可以位于5'端附近,3'端或指定长度内的任何位置。通常,接近 3' 端的序列具有更高的可信度,因此最常被优选。
3.产物Tm:熔融温度(Tm)是一半的DNA双链体解离并变成单链的温度。引物-模板DNA双链体的稳定性可以通过熔解温度(Tm).
4. 最佳退火温度(T一个选项):Rychlik 的配方最受推崇。我们的产品使用这个公式来计算它,成千上万的客户报告说,在PCR循环的退火步骤中使用它取得了良好的结果。它通常会产生良好的PCR产物产量,同时产生最少的假产物。
T一个选项 = 0.3 x(Tm引物)+ 0.7 x(Tm产品) - 14.9
其中
Tm引物是不太稳定的引物-模板对
T的熔化温度m产物是PCR产物的熔解温度。
5. 引物对Tm错配计算:引物对的两个引物应具有紧密匹配的熔解温度,以最大限度地提高PCR产物的产率。5的区别oC 或更高可导致无扩增。
使用软件进行引物设计
有许多引物设计工具可供新老用户协助PCR引物设计。这些工具可以通过降低实验失败的几率来降低实验所需的成本和时间。
Primer Premier遵循PCR引物设计的所有指南。Primer Premier可用于设计用于单个模板、比对、简并引物设计、限制性内切酶分析的引物。测序引物的重叠群分析和设计。
qPCR引物设计指南略有不同。AlleleID 和 Beacon Designer 等软件可以设计引物和寡核苷酸探针,用于复杂的检测检测,如多重检测、跨物种引物设计、物种特异性引物设计和引物设计,以降低实验成本。
PrimerPlex是一款软件,可以设计用于多重PCR和多重SNP基因分型检测的引物。